Our client, a US-based biopharmaceutical manufacturer with more than 15 years of experience in therapeutic protein production, needed to centralize terabytes of historical bioreactor cultivation data accumulated across upstream and downstream processes.
Through advanced data engineering services, Computools unified ELN, LIMS, MES, and bioreactor sensor streams into a single analytical environment. The platform enabled scalable bioreactor data analytics, improved process transparency, and established a structured foundation for predictive modeling across large-scale production systems.
The company produces therapeutic proteins using industrial-scale cell cultures grown in controlled bioreactors. Process stability directly affects purification complexity, final yield, and regulatory compliance.
To maintain competitive production performance, the organization continuously invests in bioreactor process optimization, focusing on feed strategy, oxygen control, temperature balance, and metabolic stability.
Over time, terabytes of time-series and laboratory data were generated. However, limited maturity in AI development in bioprocessing restricted the company’s ability to identify nonlinear dependencies between cultivation parameters and product quality.
The company’s production stability was affected by fragmented historical data and limited visibility across upstream and downstream processes.
Key cultivation parameters—pH, dissolved oxygen, feed strategy, and agitation—directly affected purification complexity, protein aggregation, and final yield. However, these relationships were not systematically analyzed across historical batches.
Production records were spread across ELN, LIMS, MES, and reactor systems. The lack of structured cell culture data integration created gaps in time-series alignment and slowed cross-batch comparison.
As scaling to larger bioreactors introduced additional variability, even minor parameter shifts could reduce cell viability and impact yield. Process reproducibility remained at 70–75%, and root-cause analysis often required weeks of manual consolidation. Without a unified analytical foundation, the company’s ability to evolve toward a scalable bioprocess digital twin remained constrained.
Computools designed and implemented BioreactorIQ, a centralized AI-driven analytics platform developed for industrial-scale biopharmaceutical production.
The solution unified ELN, LIMS, MES, reactor sensor streams, and laboratory measurements into a synchronized time-series architecture. Standardized parameter modeling ensured consistent comparison of pH, dissolved oxygen, feed composition, metabolic markers, and batch performance across more than 15 years of historical data.
Advanced dashboards provide full-process transparency, while machine learning models enable bioreactor predictive analytics, identifying nonlinear relationships between process parameters and product yield.
The platform also identifies optimal feed timing windows and critical cultivation phases in which parameter adjustments have the greatest impact on productivity.
The platform delivers early detection of process instability 24–48 hours before critical deviations occur and strengthens overall process control across large-scale cultivation systems. The modular architecture provides a stable foundation for long-term scalability and future expansion of the digital twin.
BioreactorIQ delivered measurable improvements across production stability and operational efficiency.
The centralized platform reduced resource waste, improved batch reliability, and generated significant annual cost savings. Onboarding time for new researchers decreased by 40–50%, ensuring faster knowledge transfer and stronger process continuity.
Improved traceability and process consistency also lowered regulatory risk and strengthened long-term production scalability.
Computools was selected for its ability to combine advanced AI engineering with the realities of large-scale biomanufacturing. The project required a structured time-series architecture, alignment of reactor and laboratory data, and predictive modeling capable of operating under production variability.
The team delivered:
Every engineering decision was aligned with measurable improvements in process stability, yield performance, and long-term scalability.
Over more than 15 years, the company accumulated terabytes of upstream and downstream production data generated across multiple bioreactor scales. While individual experiments were documented, the data remained fragmented across systems and formats, limiting cross-batch comparison and long-term process learning.
As production volumes increased, variability became more visible. Scaling from smaller to larger bioreactors introduced differences in oxygen distribution, mixing efficiency, and localized stress conditions. Differences in reactor scale and geometry led to uneven oxygen transfer and microenvironment variability, directly influencing cell growth and productivity. Even minor fluctuations in pH or dissolved oxygen can trigger metabolic shifts, reduce cell viability, and complicate downstream purification.
The absence of structured bioreactor process monitoring across historical batches made it difficult to detect patterns or anticipate instability trends.
At the same time, the company planned to expand its biologics portfolio and leverage historical R&D knowledge to better design cultivation strategies, optimize process parameters, and ensure stable product quality when bringing new biologics into production.
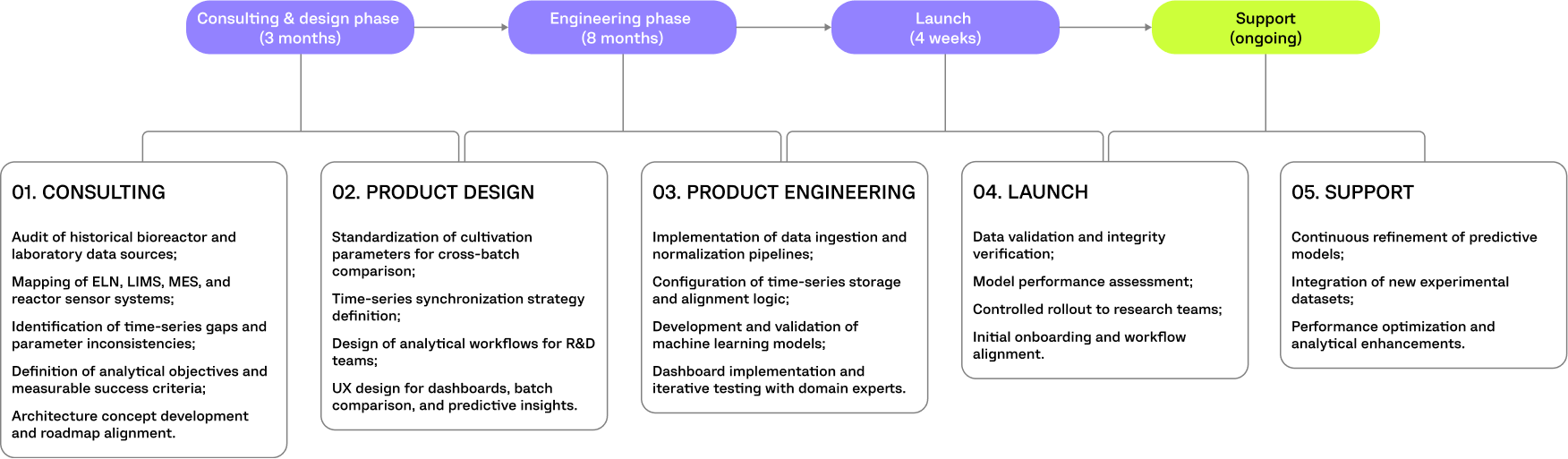
The initiative was approached as a production intelligence transformation rather than a simple data consolidation project.
The first step was to fully synchronize time-series reactor data with laboratory measurements, enabling structured upstream bioprocess optimization across feed strategies, oxygen control, temperature shifts, and metabolic markers.
The architecture was designed as a scalable biopharmaceutical manufacturing AI platform capable of processing high-volume production datasets and modeling nonlinear dependencies between cultivation conditions and downstream outcomes.
Computools served as the end-to-end technology partner, translating complex production data into a structured analytical framework.
The team led:
By connecting engineering decisions with real production workflows, Computools ensured that analytical outputs were directly applicable to operational process control.
A critical shift was the move from retrospective analysis to proactive modeling. Machine learning components were introduced to enable early cell viability prediction, identifying metabolic stress signals 24–48 hours before measurable declines.
The models incorporated early detection of lactate and ammonia accumulation as predictive stress indicators, improving the detection of viability risks before productivity losses became visible.
Embedding predictive models directly into operational dashboards allowed teams to intervene earlier, stabilize cultivation phases, and reduce downstream variability.
As the next stage of platform evolution, the client is extending the solution with a Bioprocess Digital Twin layer. This phase transforms the existing AI-driven process analytics into an interactive virtual replica of the cultivation process, enabling simulation, visualization, and predictive testing before applying changes to live bioreactors.
The planned functionality includes interactive reactor-state visualization; what-if scenario testing for feed timing, oxygen levels, and agitation strategies; predictive feedback loops that estimate the impact of yield and reproducibility; and continuous learning based on new batch and laboratory data to strengthen decision support for operators and R&D teams.
The platform design focused on translating complex bioprocess data into structured, decision-ready insights without disrupting existing production workflows. The goal was to support both R&D scientists and manufacturing engineers working with high-volume time-series datasets.

Creating a detailed profile of the primary production user to align platform design with real bioprocess workflows.
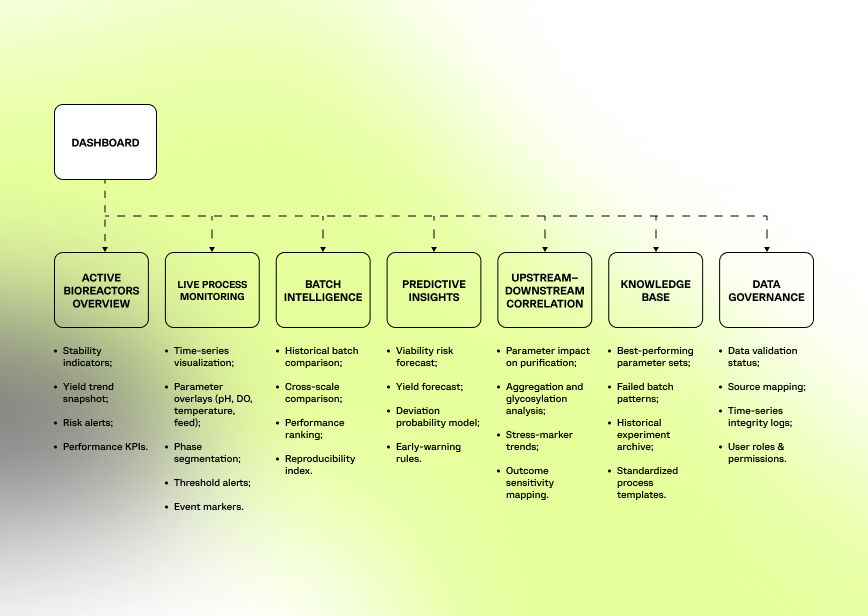
The platform structure was designed to reflect real production logic rather than abstract data categories.
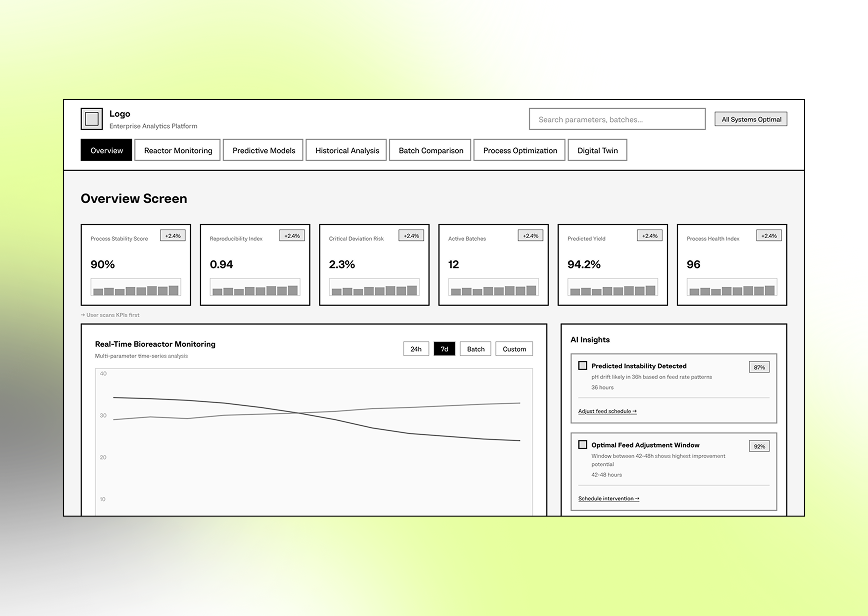
Low-fidelity layouts focused on clear visualization of time-series data, batch comparison, deviation detection, and intuitive drill-down from the process phase to specific parameter events.
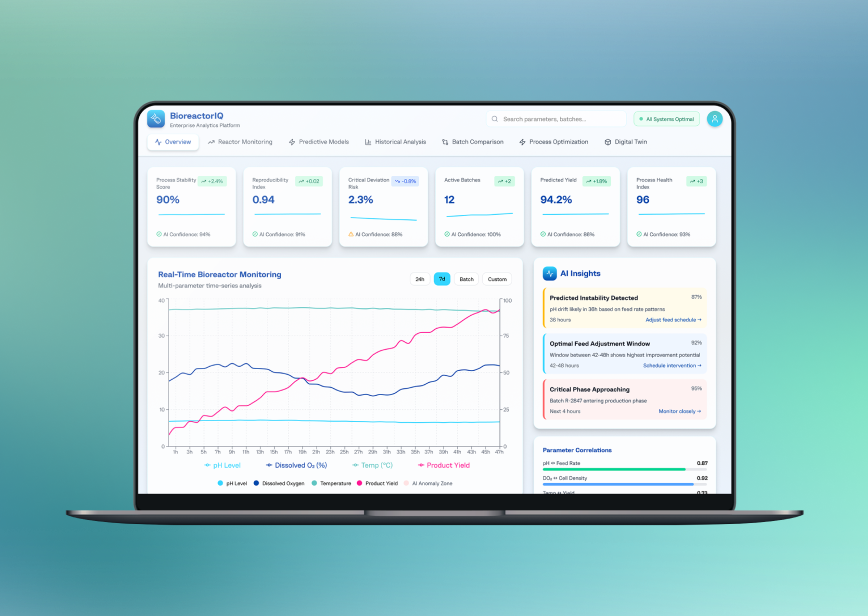
The final interface delivers interactive time-series visualization, cross-batch comparison, real-time deviation alerts, and forecast panels designed for fast, production-critical decision-making.

PYTHON
Used for data ingestion, preprocessing, feature engineering, and development of predictive models analyzing historical bioreactor experiments.

TIME-SERIES DATA STORAGE
Optimized storage layer enabling alignment of reactor sensor data (pH, DO, temperature, feed rates) with laboratory measurements across cultivation phases.

POSTGRESQL
Stores structured experiment metadata, batch configurations, and validated research results, ensuring reproducibility and traceability.

MACHINE LEARNING MODELS
Built with Python using TensorFlow and Keras, the models apply multivariate time-series architectures to identify nonlinear relationships between cultivation parameters (pH, DO, feed strategy, temperature) and downstream outcomes. They enable early detection of metabolic stress patterns, forecast drops in cell viability 24–48 hours in advance, and estimate the impact of parameter adjustments on yield and product quality.

REACT
Provides interactive dashboards for historical batch comparison, time-series visualization, and exploratory analysis of cultivation dynamics.
DOCKER
Used to containerize data processing and modeling workflows, ensuring consistent execution across research and testing environments.

TERRAFORM
Used for infrastructure-as-code provisioning, enabling reproducible deployment of cloud and hybrid environments aligned with enterprise governance policies.

SECURE INFRASTRUCTURE
Implements role-based access control, data traceability, and controlled access to sensitive experimental datasets.
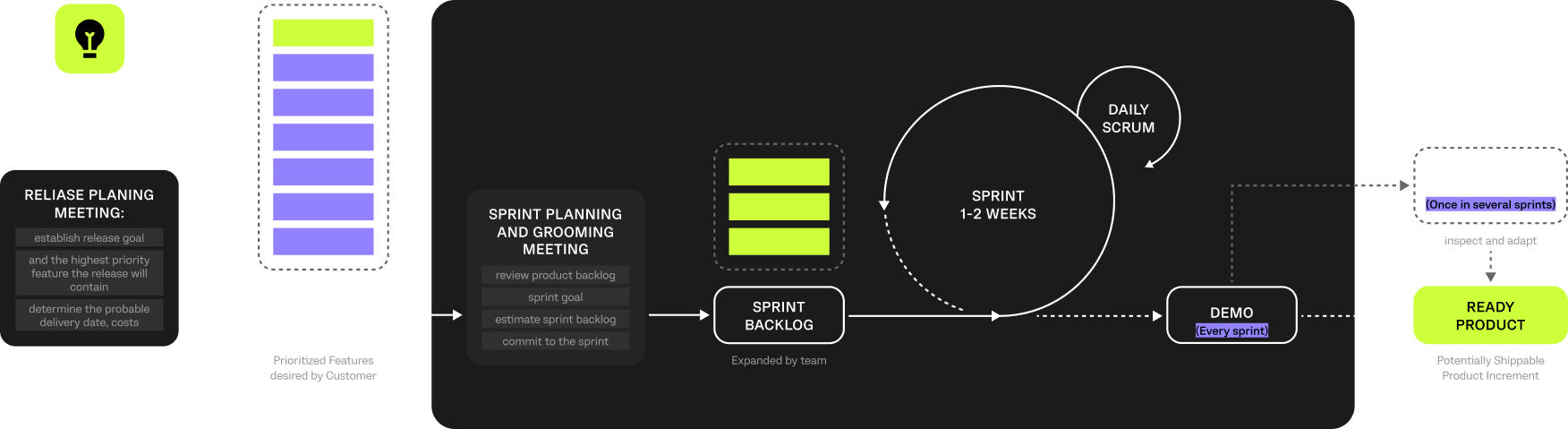
An Agile approach with Scrum was used to manage the project and support iterative development. This enabled the team to adapt to evolving research priorities, data variability, and production-specific constraints typical for bioprocess environments.
Regular sprint cycles, reviews, and stakeholder feedback ensured continuous refinement of data models, analytical logic, and predictive components, keeping the platform aligned with real R&D workflows and operational requirements.
We had years of valuable bioreactor data, but it was scattered and difficult to use effectively. Today, our team can quickly compare batches, see how process parameters interact, and understand what really drives performance. Being able to spot early warning signs before cell viability drops has changed how we manage our runs. The solution feels practical and grounded in real bioprocess work.